Recently, the research team of sugarcane stress biology both from the National Key Laboratory for Tropical Crop Biology Breeding/Institute of Tropical Bioscience and Biotechnology/Chinese Academy of Tropical Agricultural Sciences (CATAS), has made a new progress in the research on the mechanism of sugarcane response to Sporisorium scitamineum infection. The study utilized the time-ordered gene co-expression network (TO-GCN) to systematically analyze the regulatory network of sugarcane interactions with S. scitamineum, and identified two sugarcane Ca2+/H+ exchanger (ScCAX2 and ScCAX3) genes that negatively regulated plant disease resistance.

Figure 1. TO-GCN of sugarcane response to S. scitamineum stress.
(a) The process of sugarcane inoculation with S. scitamineum. (b) Relative expression values of Sspon.02G0012640-1A which was used as the initial node gene (expressed peak at 0 dpi and down-regulated at all subsequent time points) after removing two outlier samples. (c) Heat maps of average FPKMs at each level of TO-GCN at each time point after inoculation with S. scitamineum. Three different stages are identified as the un-inoculation (T0), early inoculation (T1), and late inoculation (T2), based on the expression profiles. (d) Predicted TO-GCN of sugarcane response to S. scitamineum stress. TF and non-TF genes are represented by green and pink nodes, respectively.
Sugarcane (Saccharum spp.), accounting for more than 85% of the total sugar production in China, is the most important sugar crop in the world. During growth and development, sugarcane is subject to various biotic and abiotic stresses resulting in yield reduction and quality deterioration. Sugarcane smut caused by S. scitamineum, also known as the “cancer” of sugarcane, is the most serious and widely spread fungal disease in sugarcane, causing yield losses of up to 20%-50% in severe years and even complete crop failure in extreme cases. Previous researches demonstrated that the resistance of sugarcane cultivars to smut pathogen is determined by multiple major genes and numerous micro-effector genes. Breeding resistant varieties are assumed to be the most reliable and practicable measure to prevent and control this disease. In recent years, time-course transcriptomics by TO-GCN has been conducted to obtain gene expression profiles at different time points of stress treatment, making it possible to compare multiple transcriptomes with higher accuracy and sensitivity than techniques that focus on only one time point. Time-course transcriptomics, an effective way to study gene regulatory networks, can provide valuable information to resolve the dynamic response of plants to external environmental changes, which has been widely used in wheat, cotton, poplar and other plants. However, to date, TO-GCN has not yet been reported in sugarcane. Therefore, it is of more important theoretical and practical significance to utilize the TO-GCN to construct the key metabolic networks of sugarcane response to S. scitamineum infection and to mine the key disease resistance genes.

Figure 2. Key metabolic networks of sugarcane response to S. scitamineum infection.
(a) Regulatory subnetwork of carbon metabolism pathway. (b) Regulatory subnetwork of plant hormone signal transduction pathway. (c) Regulatory subnetwork of phenylpropanoid biosynthesis pathway. (d) Regulatory subnetwork of plant pathogen interaction pathway.
In the present study, time-course transcriptome data from two different sugarcane varieties [YT93-159 (smut-resistant) and ROC22 (smut-susceptible)] post S. scitamineum infection at six time points (0 to 5 dpi) was used to construct gene co-expression networks. Besides, TO-GCN was used to explore sugarcane response to S. scitamineum treatment at time series. Potential regulatory networks of carbon metabolism, plant hormone signal transduction, phenylpropanoid biosynthesis and plant pathogen interaction pathways were then established, and the regulatory relationship between TFs and non-TFs in each subnetwork was predicted. In addition, due to the reason that Ca2+ signaling pathway was helpful to maintain cell Ca2+ stability and establish the response to sugarcane smut disease, two ScCAX genes (ScCAX2 and ScCAX3) were identified and characterized from sugarcane. They were significantly up-regulated under ABA stress but inhibited by MeJA treatment. Furthermore, overexpression of ScCAX2 and ScCAX3 enhanced the susceptibility of transgenic plants to the pathogen infection, suggesting its negative role in disease resistance. A regulatory model for ScCAX genes in disease response was thus depicted. This work helps to clarify the transcriptional regulation of sugarcane response to S. scitamineum stress and the function of the CAX gene in disease response.
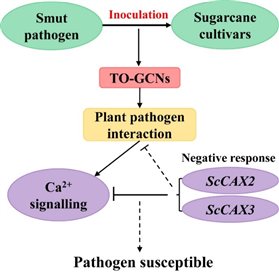
Figure 3. A regulatory model for ScCAX2 and ScCAX3 genes in disease response.
This work was already published in 《Journal of Agricultural and Food Chemistry 》under the title of 《Transcriptional regulation of sugarcane response to Sporisorium scitamineum: Insights from time-course gene co-expression and Ca2+ signaling” 》. Associate Prof. Qibin Wu from the National Key Laboratory of Tropical Crop Biology Breeding/Institute of Tropical Bioscience and Biotechnology/CATAS and Visiting Researcher of Sugarcane Research Center/CATAS Chang Zhang were the co-first authors; Prof. Youxiong Que from the National Key Laboratory of Tropical Crop Biology Breeding/Institute of Tropical Bioscience and Biotechnology/CATAS/ Fujian Agriculture and Forestry University was the corresponding author. Thanks to the funds provided by the National Key R&D Program of China, Special Projects for the Central-guided Local Science and Technology Development, China Agriculture Research System of MOF and MARA, Project of National Key Laboratory for Tropical Crop Breeding, Guangxi Key Laboratory of Sugarcane Genetic Improvement and Yunnan Key Laboratory of Sugarcane Genetic Improvement.
Article Links:https://doi.org/10.1021/acs.jafc.4c02123